Capecitabine, an antineoplastic drug used in the treatment of breast and colon cancer, can cause severe, even fatal toxicity in some patients. The interindividual variability of this toxicity is largely due to genetic variations in target genes and enzymes of metabolism of this drug, such as Thymidylate Synthase (TS) and Dihydropyrimidine Dehydrogenase (DPD). The enzyme Cytidine Deaminase (CDA), involved in the activation of capecitabine, also has several variants associated with an increased risk of toxicity to treatment, although its role as a biomarker is not yet clearly defined.
Therefore, our main objective is to study the association between the presence of genetic variants in CDA gen, CDA enzymatic activity and the development of severe toxicity in patients treated with capecitabine whose initial dose was adjusted based on the genetic profile of the DPD gen (DPYD).
MethodProspective multicenter observational cohort study, focused on the analysis of the genotype–phenotype association of the CDA enzyme.
After the experimental phase, an algorithm will be developed to determine the dose adjustment needed to reduce the risk of treatment toxicity according to CDA genotype, developing a Clinical Guide for capecitabine dosing according to genetic variants in DPYD and CDA. Based on this guide, a Bioinformatics Tool will be created to generate the pharmacotherapeutic report automatically, facilitating the implementation of pharmacogenetic advice in clinical practice. This tool will be a great support in making pharmacotherapeutic decisions based on the patient's genetic profile, incorporating precision medicine into clinical routine. Once the usefulness of this tool has been validated, it will be offered free of charge to facilitate the implementation of pharmacogenetics in hospital centers and equitably benefit all patients on capecitabine treatment.
La capecitabina, fármaco antineoplásico utilizado en el tratamiento del cáncer de mama y colon, puede dar lugar a toxicidad grave, llegando a ser mortal en algunos pacientes. La variabilidad interindividual de esta toxicidad es debida en gran medida a variaciones genéticas en genes diana y enzimas de metabolismo de este fármaco, como la Timidilato Sintasa (TS) y la Dihidropirimidina Deshidrogenasa (DPD). La enzima Citidin Desaminasa (CDA), imprescindible en la activación de capecitabina, también presenta diversas variantes asociadas con mayor riesgo de toxicidad al tratamiento, aunque su papel como biomarcador aún no está claramente definido.
Por ello, nuestro objetivo principal es estudiar la asociación entre la presencia de variantes genéticas en el gen CDA, la actividad enzimática de CDA y el desarrollo de toxicidad grave en pacientes tratados con capecitabina cuya dosis inicial se haya ajustado en base al perfil genético del gen de DPD (DPYD).
MétodoEstudio de cohortes observacional multicéntrico prospectivo, centrado en el análisis de la asociación genotipo-fenotipo de la enzima CDA.
Tras la fase experimental, se desarrollará un algoritmo que permita determinar el ajuste de dosis necesario para disminuir el riesgo de toxicidad del tratamiento en función del genotipo CDA, elaborando una Guía Clínica para la dosificación de capecitabina en función de las variantes genéticas en DPYD y CDA. En base a esta guía, se creará una Herramienta Bioinformática que genere el informe farmacoterapéutico de manera automática, facilitando la implementación del consejo farmacogenético en la práctica clínica. Esta herramienta proporcionará un gran respaldo en la toma de decisiones farmacoterapéuticas basadas en el perfil genético del paciente, incorporando la medicina de precisión en la rutina clínica. Una vez validada la utilidad de esta herramienta, se ofrecerá de manera gratuita para facilitar la implementación de la farmacogenética en los centros hospitalarios y beneficiar de forma equitativa a todos los pacientes en tratamiento con capecitabina.
Capecitabine belongs to the group of medications known as fluoropyrimidines. It is an anticancer drug used in the treatment of breast and colon cancer, and causes severe toxicity in 25.5% of treated patients1 and even death in 1.6% of such patients2. Interindividual variability in its toxic effects is partly caused by genetic variants in the enzymes involved in its metabolism and in its target genes.
The gene encoding the enzyme thymidylate synthase is a potential marker of response and toxicity to fluoropyrimidines, because the main mechanism of action of these drugs is folate cycle inhibition through the blockade of this enzyme. Several variants in this gene have been studied, and a significant association has been found between the 2R/2R genotype of the rs45445694 variant and severe toxicity to treatment (P = 0.0014; odds ratio [OR] = 5.21). Furthermore, the results of gene expression analysis in tumor tissues suggest a correlation between this genotype and low thymidylate synthase expression3.
Dihydropyrimidine dehydrogenase (DPD) is the rate-limiting enzyme in fluoropyrimidine catabolism. There is ample evidence of an association between DPD deficiency and increased levels of the active metabolite in blood, leading to severe toxicity after drug administration that can be life-threatening4. The four most relevant variants with decreased function in the gene encoding DPD (DPYD) are as follows: DPYD*2A or c.1905 + 1G > A (rs3918290); DPYD*13 or c.1679 T > G (rs55886062); c.2846A > T (rs67376798) and c.1129-5923C > G or HapB3 (rs75017182)5,6. The recommendation of the Spanish Agency of Medicines and Medical Products (AEMPS) on the dosage adjustment of fluoropyrimidines is supported by the guideline of the Clinical Pharmacogenetics Implementation Consortium7, the guideline of the Dutch Pharmacogenetics Working Group8, and the Consensus of Experts from the Spanish Pharmacogenetics and Pharmacogenomics Society and the Spanish Society of Medical Oncology9. In these guidelines, a metabolic status is established for each genotype with the corresponding recommendation supported by level I evidence (i.e. high-quality evidence).
Associations have also been found between several variants in the gene encoding the enzyme cytidine deaminase (CDA), which is essential for drug activation, and increased capecitabine toxicity. Specifically, associations have been found between the CDA variants 79 A > C (rs2072671)10–12, 451 A > G (rs532545)10,13–15, −33 ins/delC13, 92 A > G (rs602950)15, and 435 C > T (rs1048977)11 and an increased risk of treatment toxicity, with abnormal enzymatic activity being observed in some of them16.
However, despite all the published evidence, there are no clinical guidelines on its dosage according to CDA genetic variants: thus, the real prognostic value of this biomarker needs to be established through genotype–phenotype association studies. Given that almost 60% of the European population are carriers of one of these genetic variants11,16, a high percentage of the population would benefit from genetic screening for this biomarker prior to initiating treatment with capecitabine.
Regarding economic justification, a study conducted in the United Kingdom found that the average cost of consultations, hospitalization, and treatment due to the adverse effects of capecitabine was approximately €500 per patient17. These data can be extrapolated to the Spanish population, because both the ethnic group and type of healthcare model are the same. It has also been estimated that screening for DPYD and CDA genotyping in routine clinical practice could halve these costs18. Given that approximately 80 patients per year are treated with capecitabine in the participating centres, the implementation of DPYD and CDA genotyping would result in savings of about €20,000 per year per centre. If we extrapolate these data to the Spanish population as a whole, the saving would be €5,000,000 per year.
The relevance of including CDA in the dosage guideline lies in the high mutation rate of this gene. According to the dbSNP database of the National Centre for Biotechnology Information, the four most relevant DPYD variants have an average allele frequency of 0.005 in the European population, whereas the average frequency of the five selected CDA variants is 0.355. Thus, CDA screening could imply savings of 98.6% (€4,930,000) of the estimated annual savings when compared with those of the single DPYD screening currently performed, which would significantly enhance the sustainability of the Spanish Health System.
The aim of this study is to analyze the association between the presence of CDA enzyme genetic variants, its enzymatic activity, and the development of severe toxicity (grade 3–4 diarrhea, nausea/vomiting, mucositis, palmoplantar erythrodysesthesia syndrome, hepatotoxicity, and/or liver toxicity) in patients initiating treatment with capecitabine.
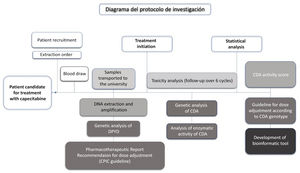
The study also includes the analysis of the four DPYD genetic variants recommended by the AEMPS: DPYD*2A or c.1905 + 1G > A (rs3918290); DPYD*13 or c.1679 T > G (rs55886062); c.2846A > T (rs67376798): and c.1129-5923C > G or HapB3 (rs75017182)19. This genotyping will be performed prior to treatment with capecitabine, allowing the initial dosage to be adjusted according to the patient's DPYD genotype. Subsequently, studies will be conducted on the allelic frequencies of the variants analyzed, the frequency and degree of toxicity among the participants, and on the dosage adjustment needed according to the CDA genetic variants for the reduction of toxicity to treatment. After the results have been validated in an independent cohort, a clinical dosage guideline for capecitabine based on DPYD and CDA genetic variants will be created and a bioinformatics tool will be developed that will match the patient's DPYD and CDA genotype with the dosage adjustment recommended in the clinical dosage guideline. Fig. 1 shows the flow chart of the different stages of the research protocol.
MethodDesignMulticentre prospective observational analytic cohort study: exposure variable = presence/absence of CDA genetic variants; response variables = severe/mild toxicity and CDA enzyme activity.
Scope of applicationThe study population will be patients initiating treatment with capecitabine at the Hospital General Universitario de Elda (Spain), Hospital General Universitario de Elche (Spain), and Hospital Virgen de los Lirios de Alcoy (Spain), during a selection period of 2 years.
Duration of the studyThe estimated total duration of the study is 4 years, divided into the following periods: selection period = 2 years; validation of the results in independent cohorts = 1 year; and the development of the Clinical Dosage Guide and the bioinformatics tool = 1 year.
Selection criteriaPatient inclusion criteria: more than 18 years of age and initiating treatment with capecitabine; giving informed consent to their participation in the study.
Patient exclusion criteria: not agreeing to participate in the study.
InterventionsThe intervention will be based on the analysis of DPYD and CDA genetic variants and the analysis of CDA enzyme activity based on routine blood tests before initiating treatment. Pharmacological treatment will based on standard clinical practice, but the DPYD polymorphism report will be taken into account to adjust the dosage prior to initiating standard treatment in patients carrying any of the risk variants.
Sample sizeSince the relative risk (RR) of developing severe toxicity in carriers of these variants remains unknown—given that previous studies have been case–control rather than cohort studies—a pilot study will first be conducted to analyze the main variables in 30 exposed patients and 30 unexposed patients. Secondly, we will calculate the sample size needed to conduct the study, taking into account the RR obtained in the pilot sample and the risk of developing the outcome in the unexposed group, while assuming, according to the published scientific evidence, a one-sided hypothesis with an α or type I error of 5% and a β οr type II error of 20% (95% confidence level and 80% power).
Given that approximately 80 patients per year are treated with capecitabine in the participating centres, and that this multicentre study includes three participating centres, approximately 240 patients could be treated per year; thus, a selection period of 2 years is considered to be of sufficient duration. If the analysis of the pilot sample shows that the sample size obtained during the selection period is too small, more centres will be invited to participate in the study, thus considerably increasing the selection rate.
VariablesThere are two main variables. The first is the incidence of severe toxicity in the exposed group and the unexposed group. Severe toxicity is considered as grade 3 or higher in relation to any of the following adverse effects classified according to CTCAE 5.0 criteria: diarrhea, nausea/vomiting, mucositis, palmoplantar erythrodysesthesia syndrome, hepatotoxicity, hematologic toxicity, and global toxicity; other types of toxicity not included among the above may be recorded. This variable is a qualitative binary variable.
The second variable is CDA enzyme activity in the exposed and unexposed groups, which will be determined after validation of the spectrophotometric method previously described by other authors16. This variable is a continuous quantitative variable, expressed as arbitrary unit (AU)/mg protein, representing micromoles of ammonia/h/mg protein.
The exposure variable is the presence of CDA genetic variants. This variable is a qualitative binary variable: carrier (homozygous mutant or heterozygous patient)/non-carrier (homozygous wild type patient). The exposed group will comprise patients carrying any of the following CDA genetic variants: 79 A > C (rs2072671); 451 A > G (rs532545); −33 ins/delC (rs3215400), 92 A > G (rs602950) and 435 C > T (rs1048977).
Study groupsThe exposed group will comprise patients carrying any of the above variants, whereas the unexposed group will comprise patients not carrying any of the variants.
Data collectionPatient selection will be conducted by the Oncology Service, and will include all patients initiating treatment with capecitabine in the participating centres and who agree to participate in the study. At the time of inclusion, and after the patient has received the information sheet and provided signed informed consent, the initial interview will be completed and a blood draw will be scheduled prior to the initiating treatment.
The prescribing oncologist will use the request form to notify the hospital's Clinical Analysis Service of the extraction date and the patient's clinical history number. After the biochemical analysis, the tube of whole blood in EDTA (2 mL) and the tube of serum (2 mL) will be stored at 4 °C and −20 °C, respectively, in the central laboratory until they are collected for genetic and enzymatic analysis.
Samples will be collected weekly and transferred to the Toxicology and Legal Medicine Laboratory at the San Juan campus of the Miguel Hernández University of Elche (Spain). This laboratory will conduct DNA extraction and purification and genotyping analysis of the four DPYD variants recommended by the AEMPS using real-time PCR with TaqMan probes in a Step One™ thermal cycler, and the results will be analyzed using StepOne v2.3 software.
The pharmacotherapeutic report including the dosage adjustment recommendation will be sent the following day by e-mail to the pharmacy service of the participating centres. This service will include the report in the patient's clinical history via the centre's computer program, allowing the corresponding oncologist to access it and initiate treatment with the appropriate regimen.
Once treatment has been initiated, the DNA samples will be frozen at −20 °C until the five CDA genetic variants have been analyzed using real-time PCR.
Serum samples will be stored at −80 °C until enzyme activity is analyzed using spectrophotometry.
Clinical treatment toxicity data will be collected on the Electronic Data Collection Sheet by the prescribing oncologist during the first six cycles of treatment according to CTCAE 5.0 criteria.
Statistical analysis will be performed after the required sample size according to the pilot sample has been reached. Using the results obtained, an association study on the genetic and enzymatic data will be conducted, and an algorithm will be developed to determine the metabolic status for each possible diplotype, including the combination of the five CDA variants and the associated toxicity risk.
After these results have been validated in an independent cohort, robust scientific evidence will be obtained to determine the needed dosage adjustment to reduce the toxicity risk in each specific genotype, thus enabling the development of a genotype-based clinical guideline for DPYD and CDA.
To facilitate the implementation of genotyping in Spanish hospitals, AlleleTyper™ software will be used to develop a bioinformatics tool that will use the algorithm. This tool will be based on matching the patient's diplotype, their metabolic status, and the dosage adjustment recommendation, thus facilitating the automatic generation of the pharmacotherapeutic report. Once the usefulness of this tool has been validated in independent cohorts, it will be offered free of charge, thereby facilitating its implementation in hospital centres to equitably benefit all patients undergoing treatment with capecitabine.
Statistical analysisA descriptive bivariate and multivariate analysis will be performed. The descriptive analysis will be performed according to the type of variant. Confidence intervals of 95% will be calculated based on the main variants (i.e. incidence of severe toxicity and CDA enzyme activity).
In the bivariate analysis, the distribution will be tested for normality: if it is normal, parametric tests will be used; otherwise, non-parametric tests will be used. Depending on the types of variables, different parametric tests will be used: Chi-square to compare two qualitative variables; Student t-test to compare a qualitative variable and a quantitative variable; and Pearson lineal correlation to compare two quantitative variables. A P-value of 0.05 will be used as cutoff for statistical significance.
In the multivariate analysis, given that the dependent variable is qualitative, binary and stepwise logistic regression will be used, calculating the RR with a 95% confidence interval. The analyses will be performed by the Statistical Studies Service of the Foundation for the Promotion of Health and Biomedical Research (FISABIO; Spain) using SPSS v.26 and R v.4.0.2 software.
DiscussionThe genotype–phenotype association project presented is novel in that, through the categorization of the CDA enzyme as a biomarker, it recommends personalized pharmacological prescription based on the DPYD and CDA genotype of each patient, rather than on DPYD alone, as is currently recommended.
After the proposed clinical guideline has been validated in an independent cohort, this approach is expected to reduce the risk of toxicity of capecitabine treatment due to overdosage in the case of patients carrying ultrarapidly metabolizing CDA genetic variants and/or in those carrying poorly metabolizing DPYD genetic variants.
One of the main differences between this project and those already published lies in its prospective cohort design. The great majority of studies are retrospective, are more limited, and only associate polymorphisms with toxicity, without performing dosage adjustments11,20,21. The present study will prospectively perform determinations and pharmacogenetic counseling, by taking advantage of the resources and clinical benefit of performing preventive genotyping. In addition, statistical analysis will provide us with the RR of the presence of CDA variants, which is a more powerful statistical measure than those obtained in case–control studies. It should also be noted that, thanks to the development of the bioinformatics tool, it will be possible to implement the Pharmacogenetic Analysis Service, thus facilitating the interpretation of the results in the clinical practice of hospital centres.
Finally, the categorization of CDA as a biomarker could also be useful in other drug treatments, given that this enzyme is involved in the inactivation of gemcitabine and cytarabine, which are antineoplastic drugs used for the treatment of various types of cancer. Thus, the results obtained in this project could be used in subsequent studies with the aim of developing clinical dosage guidelines for such drugs and reducing chemoresistance to treatment and consequent disease progression.
LimitationsThe fact that the metabolic phenotype is established by enzymatic activity alone may be a limitation of this study, given that it is also relevant to know each patient's pharmacokinetics in relation to their capecitabine metabolism in order to describe a more complete phenotype. This aspect could be a future research line, extending the characterization of CDA as a biomarker to the phenotype by including pharmacokinetic analysis during the period in which each patient receives their first dosage of capecitabine. In this regard, a population pharmacokinetic model has recently been used to study the relationship between the metabolites of capecitabine (5-FU, 5’-DFCR, and 5’-DFUR) and toxicity or clinical response to the drug22. The data obtained identify the CDA enzyme as a statistically significant covariate, thus highlighting the usefulness of simultaneously analyzing genetic, enzymatic, and pharmacokinetic data for this biomarker.
Ethical responsibilitiesThis project has been classified by the Spanish Agency of Medicines and Medical Products (AEMPS), with code ATB-CAP-2020-01, as a “Post-authorization prospective follow-up study” (EPA-SP). It has been authorized by the Autonomous Committee for the Evaluation of Observational Post-authorization Studies, for Prospective Follow-up Studies with Medicines (CAEPO). It has also been approved by the Drug Research Ethics Committee (CEIM) of the General University Hospital of Elda (Spain) and the General University Hospital of Elche (Spain) in accordance with current legislation: Royal Decree 223/2004 of February 6, 2004.
Funding- V Call for Grants for the Promotion of Research Activity of the Foundation for the Promotion of Health and Biomedical Research of the Community of Valencia, Spain (FISABIO) 2021.
- Grants for Preparatory Actions to support the exploration and formulation of future joint research and innovation projects between the Universidad Miguel Hernández de Elche (Spain) and FISABIO 2021.
- VIII Call for grants for the Support and Promotion of Research of the Instituto de Investigación Sanitaria y Biomédica de Alicante, Spain (ISABIAL) 2021, modality B.
- Roche sponsorship with code CPRES00254.
The following authors contributed to the development of the article and its preparation:
Paula Castro-Sánchez: promotion of the study, development of the protocol, request for classification and authorizations needed for its approval in the participating centres, general coordination of the study, and drafting the article.
Andrés Corno-Caparrós: design, organization, and execution of the genetic analysis of the study, planning and development of the bioinformatics tool, critical review of the article, and approval of the final version for publication.
M. Amparo Talens-Bolós: design and organization of sample logistics between the participating hospitals and the university, issuing the pharmacogenetic reports, critical review of the article, and approval of the final version for publication.
María José Prieto-Castelló: application for public funding for the study, data management for statistical analysis, critical review of the article, and approval of the final version for publication.
Loreto Pitaluga-Poveda: validation of the enzyme activity analysis method, management and preparation of reagents, critical review of the article, and approval of the final version for publication.
Juan Antonio Barrera-Ramírez: organizing and conducting patient recruitment and clinical data collection, critical review of the article, and approval of the final version for publication.
We would like to thank all the professional at the hospital pharmacy, medical oncology, and clinical analysis departments of the participating centres who collaborated in the development of this project. We would also like to thank the ANCOR Genetic Analysis Laboratory for its technical and intellectual support, which facilitated the development of this study.